Content-based image retrieval (short CBIR) has been around for more than three decades. Before 2012, when deep learning achieved its first major successes, CBIR methods were rather primitive mainly due to the “semantic gap”; what algorithms found to be similar did not conform to the physicians’ perception. Conventional (non-AI) features created the semantic gap: color, shape, and texture attributes of digital images that were neither salient nor invariant to imaging parameters. Deep learning changed all that.

A pre-trained neural network of sufficient depth can be employed as a feature extractor; the so-called deep embeddings (generally last filtering layers in a pre-trained convolutional network) can represent images with unprecedented semantic conformance. Such robust representations can be used to tags, i.e., index, histopathology images for search and retrieval purposes. A large number of AI-based CBIR methods have been proposed in recent years. There is no doubt that we have the technology to implement CBIR systems for clinical utility. But will we?
That we can do CBIR does not mean we will. There are still several major prerequisites to be fulfilled before pathology departments can install and use CBIR systems with tangible contributions toward more accurate diagnosis and treatment planning. DICOM and PACS, VNA or not, in digital pathology are, compared to the radiology, in their infancy. The design, validation, and deployment of capable CBIR solutions are virtually impossible if there are still no widely adopted rules, best practices, and specifications for acquiring, storing, and accessing whole slide images. In addition, communication protocols are indispensable for interoperability; we experienced that during the 90s when radiology went digital. Getting rid of the physical archives of millions of glass slides has always been an argument for going digital. However, we know that the gigapixel nature of whole slide images may shrink the physical space necessary for digital storage, but it comes at high upfront expenses to purchase high-performance storage, a factor that the individual pathologist may not perceive as an obstacle as this is mainly a headache for the CFOs and CEOs of the clinics and hospitals. Needless to say that the true potential of CBIR cannot be possibly explored unless continuous access to large archives of histopathology images is established and available at all times. Assuming that standards for interoperability are enforced, and high-performance devices are available, CBIR may still not be feasible for clinical utility. The so-called “indexing” of whole slide images is not a cheap operation. Generation of those salient deep features that close the semantic gap comes at a cost: one needs massive GPU power to perform the indexing (computing deep features). With a capable GPU, you may need several minutes per whole slide image depending on the size of the tissue sample, and desired magnification. The indexing of an archive with 100,000 whole slide images, which may be the data of just several thousand patents, may need 20 weeks of constant undisrupted operation of multiple GPUs. The indexing is supposed to be a one-time process: you only index once. But for experimental and not thoroughly validated software, the indexing may take several attempts to reliably tag all images in a large archive. The issues with clinical utility do not end at DICOM, VNA, storage, and GPUs. The main requirement for CBIR is the existence of an archive where metadata is associated with the whole slide images.
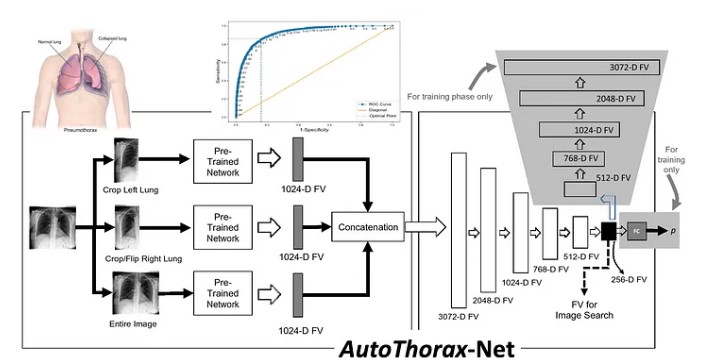
Whereas supervised AI (classification, segmentation, detection, and prediction) need a lot of well-delineated (labeled) data to be trained before clinical deployment, unsupervised AI (clustering, search, and visualization) do need the data after the deployment; yes, images do not need to be labeled, but it does need the associate metadata: diagnostic reports, genome, and treatment outcomes. Searching and retrieving similar tissue morphologies may be a challenging task and hence applaudable, but it isn’t very helpful if it is not accompanied by the corresponding evidence for the past cases. Where are these metadata? Where are diagnostic reports, RNA sequences, and patient outcomes stored? Is that in image management systems or in lab information systems? Do we need to access RIS and HIS as well? CBIR won’t provide any value unless we also retrieve the evidence for recommending a similar diagnosis or treatment.
CBIR is merely a facilitator; its value is to assist the pathologist in accessing the accumulated knowledge in evidently diagnosed and treated past cases. Do all these challenges and obstacles mean we may not be able to see a post-pandemic boom of AI-powered CBIR systems for clinical use within the next few years? Most certainly not. The low-hanging CBIR fruit is in small but well-curated datasets for each primary diagnosis. Recently published results have shown that the majority vote among retrieved cases through CBIR can reach acceptable and reliable accuracy levels for as little as several hundred samples (i.e., cases) per primary diagnosis. Creating representative “atlases” of images with their metadata can undoubtedly be done in a short time without being at the mercy of a solution for all the hindrances mentioned above. Internal virtual consultations through CBIR are certainly possible. It won’t be the universal solution that CBIR can be, but it’s the low-hanging fruit that we can harvest to bridge the gap toward wider interoperability in digital pathology, or a national, or even better, an international archive of millions of cases.
First version published on 08/23/2021 on the website of Digital Pathology Association.