This page offers code and data related to the paper
“Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides” by Abtin Riasatian et al.

The adoption of digital pathology that replaces the microscope with a digital scanner and computer monitor has gained momentum in recent years. The process of digitization of whole slide images (WSIs) offers many advantages such as more efficient workflows, easier collaboration and telepathology, and new biological insights into histopathology data through the usage of image processing and computer vision algorithms to detect relevant clinicopathologic patterns. But image analysis in pathology, especially compact and expressive image representation as a fundamental task in this area, has its own challenges, such as the large size of WSIs, texture complexity, and polymorphism. Handcrafted features, i.e., image descriptors that have been manually designed based on general image processing knowledge, have been in use as a solution for several decades. However, with the recent progress in machine learning, particularly deep learning, studies have shown that deep features, i.e., high-level embeddings in a properly trained deep network, can outperform handcrafted features in most applications and are considered the most robust and expressive source for image representation.
As a result, many different convolutional architectures have been trained and introduced to provide features either directly or through transfer learning. Pre-trained networks such as DenseNet that draw their discrimination power from intensive training with millions of natural (non-medical) images have found widespread usage in medical image analysis. In addition, several attempts have been reported in the literature to fine-tune or train deep networks with histopathology images to improve the representation power of the deep features, a desirable task that is impeded by the lack of labeled image data and the need for high-performance computing devices. Also, while it is generally expected that fine-tuning and training should deliver more accurate results than transfer learning, the data and experimental challenges inherent in this may easily prevent the expected effects. So this has created new questions in medical image analysis as to which network topology is most suitable for a given task. Is transfer learning sufficient to solve specific problems? What challenges and benefits does training an entire network from scratch entails?
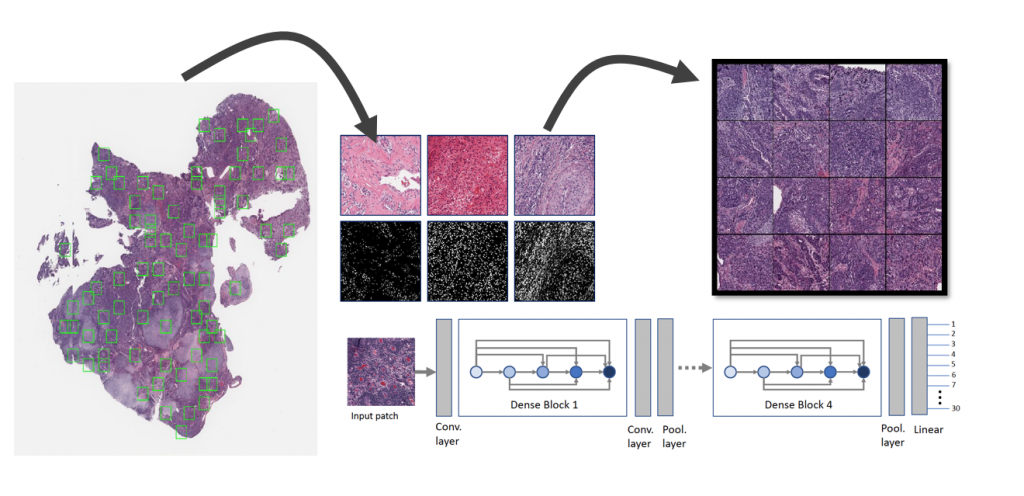
extracted from TCGA whole slide images.
In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000 × 1000 pixels acquired at 20× magnification through our proposed “high-cellularity mosaic” approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch normalized ReLU (CBR) networks.
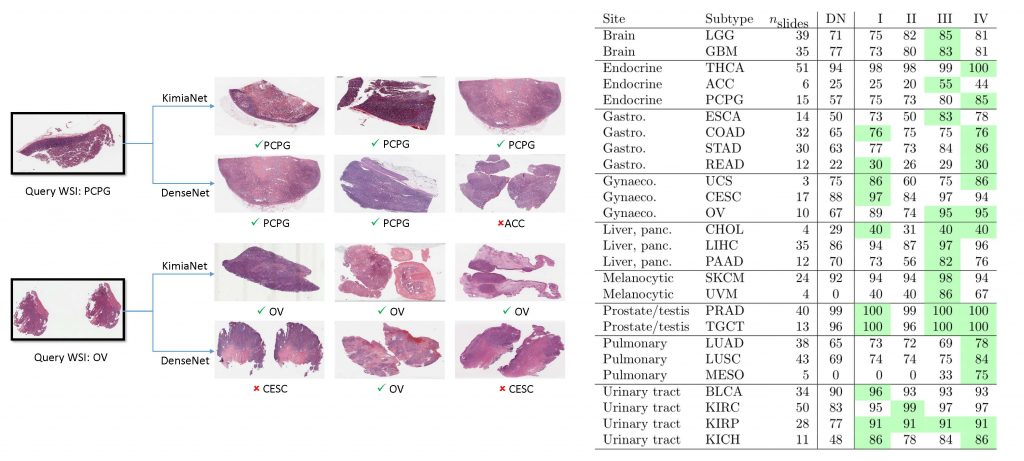
The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as a feature extractor to represent histopathology images. The main contribution of this work is exploiting a diverse, multi-organ public image repository like TCGA at 20× magnification to extract large patches, 1000×1000 pixels at high resolution, for training a densely connected network with weak labels to serve as feature extractor utilizing the proposed high-cellularity mosaic approach.
To Download Code/Data visit: https://github.com/RhazesLab/KimiaNet