Yottixel is a proposed search engine for digital histopathology archives, we report the results from searching the largest public repository (The Cancer Genome Atlas, TCGA) of whole-slide images. We successfully indexed and searched almost 30,000 high-resolution digitized slides. The proposed method achieved high accuracy values +90% for many cancer types. ” Pan-cancer diagnostic consensus through searching archival histopathology images using artificial intelligence” by Shivam Kalra et al.

A system that combines artificial intelligence (AI) with human knowledge promises faster and more accurate cancer diagnosis. Yottixel is a powerful image search engine, developed by a team led by researchers at Kimia Lab (University of Waterloo). Yottixel uses digital images of tissue samples to match new cases of suspected cancer with previously diagnosed cases in a database within a fraction of a second. The search engine is validated using a largest publicly available archive in the world – comprised of about 30,000 digitized slides from almost 11,000 patients – the technology achieved high accuracy values (in some cases high 90s) for 32 forms of cancer in 25 organs and body parts.
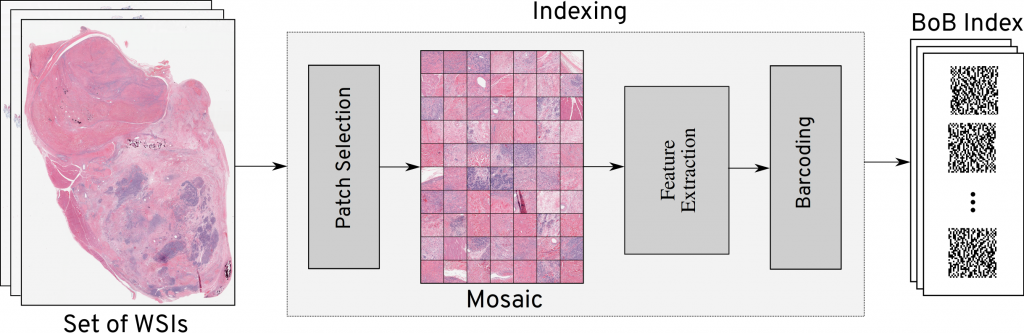
The major novelty of the Yottixel is the approach it uses for representing whole slide images (WSIs). Each WSI is converted to a set of representative patches that are converted into barcodes using deep models. These barcodes, called Bunch of Barcodes (BoB) are a compact form of characterization of a WSI. The BoB index requires less computation and storage resources for searching in large archives of histopathology slides compared to other approaches. For example, a WSI of size 200-300 MB can be converted to a BoB index of ~10 KB offering up to 99.9% reduction in its size.
Note: This technology is patented (see patent 1, patent 2). All commercial rights are owned exclusively by Huron Digital Pathology.
To Download Code/Data visit: https://github.com/RhazesLab/yottixel