Aug 2018
Whole-slide imaging (WSI) technology has emerged as a powerful tool in pathology analysis, revolutionizing the identification and understanding of diseases. Unlike traditional pathology analysis, which has limitations in assessing tissue samples, WSI allows for high-resolution digital scanning of samples, enabling easy sharing and collaboration among multiple pathologists. However, the visual inspection of detailed WSI scans can be time-consuming and inefficient. To address this challenge, the development of computer-aided diagnosis (CAD) systems becomes crucial in improving the efficiency and reliability of digital pathology analysis.
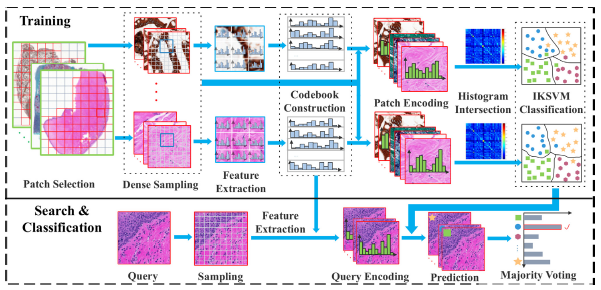
Computer-aided diagnosis (CAD) systems have played a significant role in digital pathology, focusing on tasks such as cell detection, segmentation, retrieval, and classification. These systems rely on various features, including texture and color information, to detect and segment cells. Techniques such as the scale-invariant feature transform (SIFT) and local binary patterns (LBP) are commonly used for search and classification of pathology images. However, imbalanced training datasets can introduce biases, as the most frequent texture samples tend to dominate the codebook formation.
To overcome the limitations of biased codebooks caused by imbalanced training samples, a histopathology image retrieval framework utilizing multiple bag-of-words (MBoW) approach is proposed. This framework creates multiple disjoint dictionaries for each class and leverages the histogram intersection kernel SVM (IKSVM) for classification. By employing multiple dictionaries and avoiding dominance of certain texture types, this approach aims to build more expressive and discriminative dictionaries.
The proposed framework is validated using the Kimia Path24 image dataset, consisting of 24 WSI scans with different tissue textures. The dataset includes thousands of patches, with a subset designated for testing. The main contribution of the study lies in the design of the “multiple BoW” framework with disjoint training, effectively mitigating the biased codebook problem. Experimental results demonstrate the superiority of the MBoW approach over other methods, achieving a state-of-the-art total accuracy of 76.09% for the Kimia Path24 dataset. These results underline the significance of accurate retrieval algorithms in medical imaging, where the prior knowledge of class labels is often available.
The study presents a comprehensive literature review on image classification and retrieval in digital pathology, the bag-of-words methodology, and the local binary patterns descriptor. It sets the stage for the proposed research, introducing the concept of multiple dictionaries and their advantages. The experimental results highlight the effectiveness of the MBoW approach in overcoming biased codebooks and improving retrieval accuracy. The proposed framework holds promise in enhancing the efficiency and reliability of digital pathology analysis, ultimately leading to better disease identification and understanding.
In conclusion, the application of multiple bag-of-words approach in whole-slide imaging (WSI) technology presents a significant advancement in pathology analysis. By addressing the challenges associated with biased codebooks and leveraging the histogram intersection kernel SVM, the proposed framework improves retrieval accuracy and offers promising results. The integration of accurate retrieval algorithms in medical imaging holds great potential for enhancing diagnostic capabilities and furthering our understanding of diseases.
Additional details: Multiple disjoint dictionaries for representation of histopathology images