March 2021
Accessing and utilizing large archives of digital pathology scans pose significant challenges due to the limitations of current text-based search methods. Traditional content-based image retrieval (CBIR) approaches are insufficient for the complex and variable nature of pathology images. This article introduces Yottixel, a novel approach that leverages deep learning architectures and intelligent indexing techniques to improve the efficiency and effectiveness of searching in digital pathology archives. By addressing the limitations of existing CBIR systems and integrating computer-assisted diagnosis (CAD) systems, Yottixel aims to revolutionize pathology routines. Previous Works in Histopathology Image Retrieval: The article reviews previous works related to histopathology image retrieval, highlighting the importance of integrating CAD systems into pathology routines. It discusses the limitations of existing CBIR systems, such as their reliance on basic image features and inability to handle large image dimensions and variability. These limitations emphasize the need for a more advanced and efficient approach, leading to the development of Yottixel.
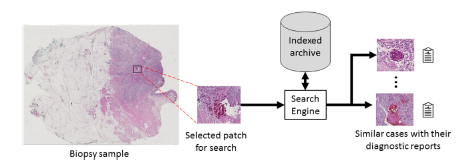
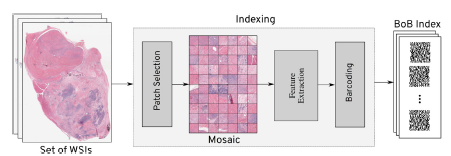
Design and Implementation: Yottixel is a comprehensive image retrieval and indexing framework specifically designed for whole slide images (WSIs) in computer-aided diagnosis systems. It comprises two main contributions: a mosaic-based representation of WSIs and an ensemble framework for indexing and retrieving WSIs using barcodes. The indexing process in Yottixel involves two phases: offline indexing and runtime search. During offline indexing, computational resources are used to index available WSI files, preferably on scanners with GPU power. Once a sufficient number of images are indexed, both phases are activated simultaneously, with runtime search given higher priority. This approach ensures optimal utilization of resources in laboratory and hospital settings. The core component of Yottixel is its indexing structure, which determines the speed and reliability of search results. Yottixel indexes a WSI by computing its mosaic, a representative set of patches, and converting it into a “Bunch of Barcodes” (BoB) index. This indexing algorithm accounts for the large number of WSI files generated annually in pathology laboratories, optimizing efficiency despite limited computing and storage infrastructure. The mosaic creation algorithm segments a WSI into different regions based on color composition using the k-means algorithm. By randomly selecting a small percentage of patches from the color-segmented regions while preserving spatial diversity, Yottixel ensures a diverse representation of patches. The resulting mosaic, typically 20 times smaller than the original WSI, captures pattern variability from a computer vision perspective. The mosaic patches are then converted into a set of barcodes, serving as the index for each WSI. Feature vectors are extracted using the DenseNet architecture, pretrained on natural images from the ImageNet dataset. The feature vectors undergo Global Average Pooling to obtain a 1024-dimensional vector, which is binarized using the discrete differentiation or MinMax algorithm, resulting in a binary barcode representation. This barcode enables fast Hamming distance search and significantly reduces storage requirements. Yottixel Search Modes and Validation: Yottixel supports two search modes: vertical and horizontal. In the vertical mode, image matching is limited to the same primary site as the query patch/WSI for all patients. In the horizontal mode, the entire index is searched across all primary sites for all patients. Users can interact with Yottixel through an interactive interface to perform search queries on their WSIs. To validate Yottixel’s search capabilities, two datasets were used. The first dataset, provided by the University of Pittsburgh Medical Center (UPMC), consisted of 300 H&E stained WSIs representing over 80 different primary diagnoses from various organs. The second dataset was obtained from The Cancer Genome Atlas (TCGA) and included 2,020 WSIs from a collection of over 33,000 WSIs. Both datasets contained WSIs with FFPE tissue specimens.
The experiments evaluated the search engine’s performance in terms of retrieval accuracy and classification capability. A “leave-one-out” strategy was employed for cross-validation, measuring search accuracy using hit rates and the number of correct retrievals. Results indicated that the search engine outperformed random retrieval, particularly for primary diagnosis matching. The impact of mosaic size on search accuracy was also analyzed, demonstrating that smaller mosaic subsets can achieve comparable accuracy to larger mosaics, especially for primary diagnosis matching. The search engine’s classification capability was evaluated by performing vertical searches within the same primary site. High recall values for certain primary sites indicated Yottixel’s ability to distinguish between different tumor types within those sites. User feedback from both expert pathologists and non-expert users affirmed the effectiveness of Yottixel’s search results, with higher-ranking results being more positively ranked.
The experiments demonstrated the feasibility and efficacy of Yottixel as a search engine for digital pathology archives. By overcoming the limitations of existing CBIR systems and incorporating advanced deep learning architectures and intelligent indexing techniques, Yottixel provides more accurate and efficient retrieval of relevant pathology images. The results validate the need for further investigation and improvement, making Yottixel a valuable tool for diagnostics, research, and education in the field of digital pathology.
Additional details: Yottixel – An Image Search Engine for Large Archives of Histopathology Whole Slide Images